dl_binder_design - protein design via deep learning : 2024-11-01 dl_binder_design The Baker Lab presents a new method that combines deep learning and physics-based approaches to design high-affinity protein binders. The method increases design success rates by a factor of ten .
dl_binder_design10 AWESOME AFFORDABLE ALTERNATIVES TO THE CARTIER TANK!!! Featuring a super cool new Casio, microbrand alternatives and a classy Hamilton. Buy .
$18.71
dl_binder_designLearn how to design binders based on scaffolds using three environments: SE3nv, dl_binder_design and af2_binder_design. See the scripts, parameters and directories for each environment and the evaluation of .This article explores the use of deep learning methods to improve the success rate of designing high affinity protein binding proteins from target structural information alone. . The Baker Lab presents a new method that combines deep learning and physics-based approaches to design high-affinity protein binders. The method increases design success rates by a factor of ten .
dl_binder_design We use a pseuodocycle-based shape complementarity optimizing approach to design nanomolar binders to diverse ligands, including the flexible and polar .
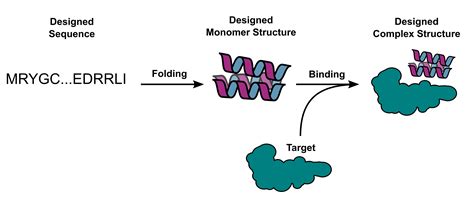
17 Citations. 226 Altmetric. Metrics. Many peptide hormones form an α-helix on binding their receptors 1, 2, 3, 4, and sensitive methods for their detection could . Here, we explore the augmentation of energy-based protein binder design using deep learning. We find that using AlphaFold2 or RoseTTAFold to assess the . It is often desirable to be able to specify a protein fold during design (such as triose-phosphate isomerase (TIM) barrels or cavity-containing NTF2s for small molecule binder and enzyme design 32 .
Posted: 4 April 2023 8:16 am. News. amaysim launches 4 international roaming packs to use in 90+ countries. Budget provider amaysim has replaced its existing 'Pay As You .
dl_binder_design